AtLOV2
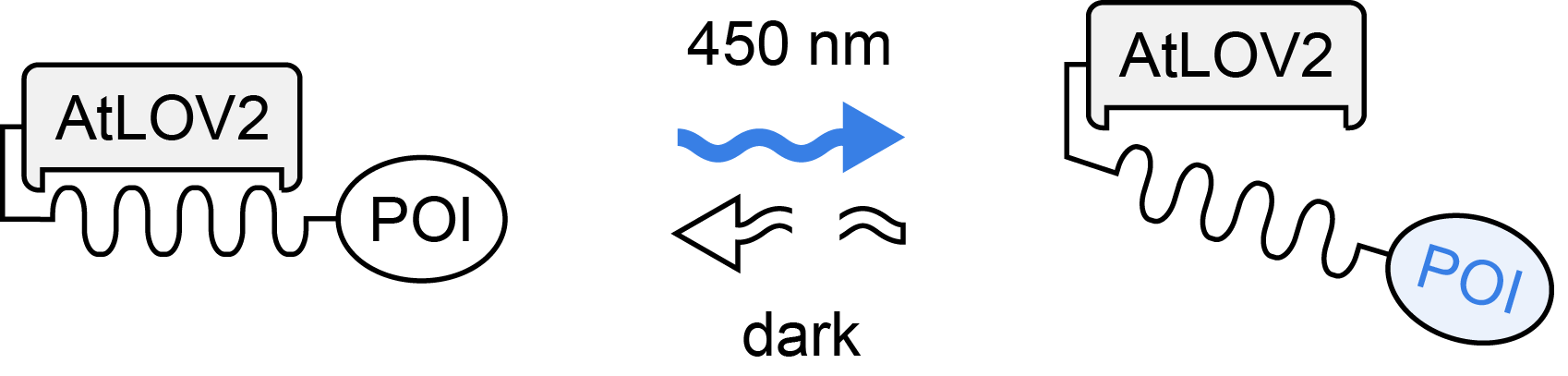
| Photoreceptor | AtLOV2 |
| Binding partner | / |
| Cofactor | FMN |
| Source organism | Arabidopsis thaliana |
| Mode of action | intramolecular conformational change |
| Excitation wavelength | 450 nm |
| Reversion wavelength | dark |
| Excitation time | seconds |
| Reversion time | tens of seconds |