CcaS/CcaR
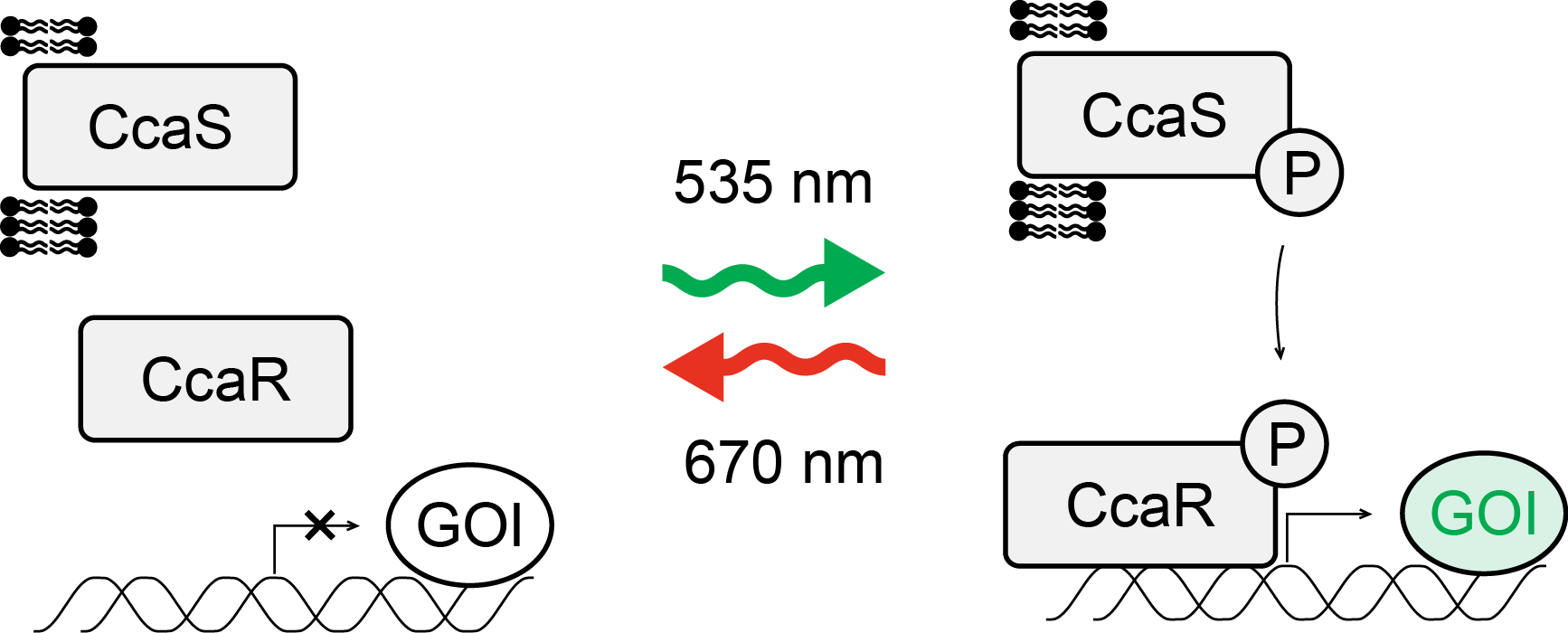
| Photoreceptor | CcaS |
| Binding partner | CcaR |
| Cofactor | PCB |
| Source organism | Synechocystis sp. PCC 6803 |
| Mode of action | gene expression |
| Excitation wavelength | 535 nm |
| Reversion wavelength | 670 nm |
| Excitation time | ? Add information |
| Reversion time | ? Add information |